Marius George Linguraru
Standardized Methods and Recommendations for Green Federated Learning
Jan 30, 2026Abstract:Federated learning (FL) enables collaborative model training over privacy-sensitive, distributed data, but its environmental impact is difficult to compare across studies due to inconsistent measurement boundaries and heterogeneous reporting. We present a practical carbon-accounting methodology for FL CO2e tracking using NVIDIA NVFlare and CodeCarbon for explicit, phase-aware tasks (initialization, per-round training, evaluation, and idle/coordination). To capture non-compute effects, we additionally estimate communication emissions from transmitted model-update sizes under a network-configurable energy model. We validate the proposed approach on two representative workloads: CIFAR-10 image classification and retinal optic disk segmentation. In CIFAR-10, controlled client-efficiency scenarios show that system-level slowdowns and coordination effects can contribute meaningfully to carbon footprint under an otherwise fixed FL protocol, increasing total CO2e by 8.34x (medium) and 21.73x (low) relative to the high-efficiency baseline. In retinal segmentation, swapping GPU tiers (H100 vs.\ V100) yields a consistent 1.7x runtime gap (290 vs. 503 minutes) while producing non-uniform changes in total energy and CO2e across sites, underscoring the need for per-site and per-round reporting. Overall, our results support a standardized carbon accounting method that acts as a prerequisite for reproducible 'green' FL evaluation. Our code is available at https://github.com/Pediatric-Accelerated-Intelligence-Lab/carbon_footprint.
FeTTL: Federated Template and Task Learning for Multi-Institutional Medical Imaging
Jan 22, 2026Abstract:Federated learning enables collaborative model training across geographically distributed medical centers while preserving data privacy. However, domain shifts and heterogeneity in data often lead to a degradation in model performance. Medical imaging applications are particularly affected by variations in acquisition protocols, scanner types, and patient populations. To address these issues, we introduce Federated Template and Task Learning (FeTTL), a novel framework designed to harmonize multi-institutional medical imaging data in federated environments. FeTTL learns a global template together with a task model to align data distributions among clients. We evaluated FeTTL on two challenging and diverse multi-institutional medical imaging tasks: retinal fundus optical disc segmentation and histopathological metastasis classification. Experimental results show that FeTTL significantly outperforms the state-of-the-art federated learning baselines (p-values <0.002) for optical disc segmentation and classification of metastases from multi-institutional data. Our experiments further highlight the importance of jointly learning the template and the task. These findings suggest that FeTTL offers a principled and extensible solution for mitigating distribution shifts in federated learning, supporting robust model deployment in real-world, multi-institutional environments.
MRI-to-CT Synthesis With Cranial Suture Segmentations Using A Variational Autoencoder Framework
Dec 29, 2025Abstract:Quantifying normative pediatric cranial development and suture ossification is crucial for diagnosing and treating growth-related cephalic disorders. Computed tomography (CT) is widely used to evaluate cranial and sutural deformities; however, its ionizing radiation is contraindicated in children without significant abnormalities. Magnetic resonance imaging (MRI) offers radiation free scans with superior soft tissue contrast, but unlike CT, MRI cannot elucidate cranial sutures, estimate skull bone density, or assess cranial vault growth. This study proposes a deep learning driven pipeline for transforming T1 weighted MRIs of children aged 0.2 to 2 years into synthetic CTs (sCTs), predicting detailed cranial bone segmentation, generating suture probability heatmaps, and deriving direct suture segmentation from the heatmaps. With our in-house pediatric data, sCTs achieved 99% structural similarity and a Frechet inception distance of 1.01 relative to real CTs. Skull segmentation attained an average Dice coefficient of 85% across seven cranial bones, and sutures achieved 80% Dice. Equivalence of skull and suture segmentation between sCTs and real CTs was confirmed using two one sided tests (TOST p < 0.05). To our knowledge, this is the first pediatric cranial CT synthesis framework to enable suture segmentation on sCTs derived from MRI, despite MRI's limited depiction of bone and sutures. By combining robust, domain specific variational autoencoders, our method generates perceptually indistinguishable cranial sCTs from routine pediatric MRIs, bridging critical gaps in non invasive cranial evaluation.
BabyFlow: 3D modeling of realistic and expressive infant faces
Dec 22, 2025Abstract:Early detection of developmental disorders can be aided by analyzing infant craniofacial morphology, but modeling infant faces is challenging due to limited data and frequent spontaneous expressions. We introduce BabyFlow, a generative AI model that disentangles facial identity and expression, enabling independent control over both. Using normalizing flows, BabyFlow learns flexible, probabilistic representations that capture the complex, non-linear variability of expressive infant faces without restrictive linear assumptions. To address scarce and uncontrolled expressive data, we perform cross-age expression transfer, adapting expressions from adult 3D scans to enrich infant datasets with realistic and systematic expressive variants. As a result, BabyFlow improves 3D reconstruction accuracy, particularly in highly expressive regions such as the mouth, eyes, and nose, and supports synthesis and modification of infant expressions while preserving identity. Additionally, by integrating with diffusion models, BabyFlow generates high-fidelity 2D infant images with consistent 3D geometry, providing powerful tools for data augmentation and early facial analysis.
Adaptable Segmentation Pipeline for Diverse Brain Tumors with Radiomic-guided Subtyping and Lesion-Wise Model Ensemble
Dec 16, 2025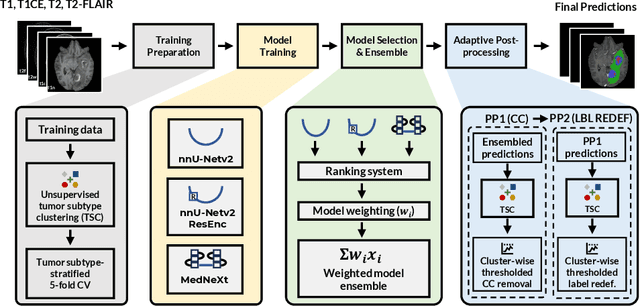
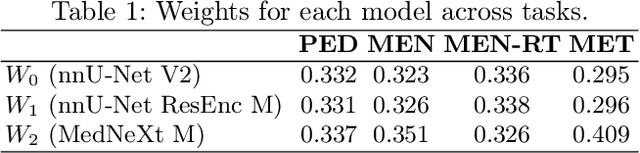
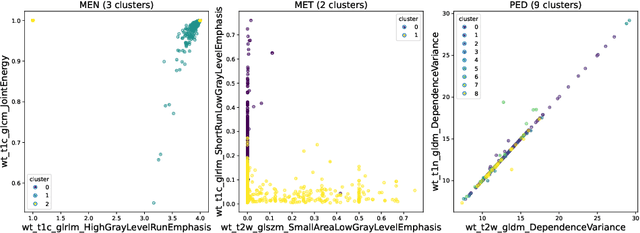
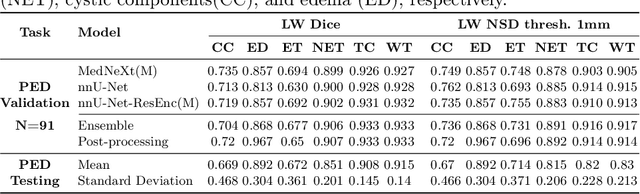
Abstract:Robust and generalizable segmentation of brain tumors on multi-parametric magnetic resonance imaging (MRI) remains difficult because tumor types differ widely. The BraTS 2025 Lighthouse Challenge benchmarks segmentation methods on diverse high-quality datasets of adult and pediatric tumors: multi-consortium international pediatric brain tumor segmentation (PED), preoperative meningioma tumor segmentation (MEN), meningioma radiotherapy segmentation (MEN-RT), and segmentation of pre- and post-treatment brain metastases (MET). We present a flexible, modular, and adaptable pipeline that improves segmentation performance by selecting and combining state-of-the-art models and applying tumor- and lesion-specific processing before and after training. Radiomic features extracted from MRI help detect tumor subtype, ensuring a more balanced training. Custom lesion-level performance metrics determine the influence of each model in the ensemble and optimize post-processing that further refines the predictions, enabling the workflow to tailor every step to each case. On the BraTS testing sets, our pipeline achieved performance comparable to top-ranked algorithms across multiple challenges. These findings confirm that custom lesion-aware processing and model selection yield robust segmentations yet without locking the method to a specific network architecture. Our method has the potential for quantitative tumor measurement in clinical practice, supporting diagnosis and prognosis.
Improving Pre-trained Segmentation Models using Post-Processing
Dec 16, 2025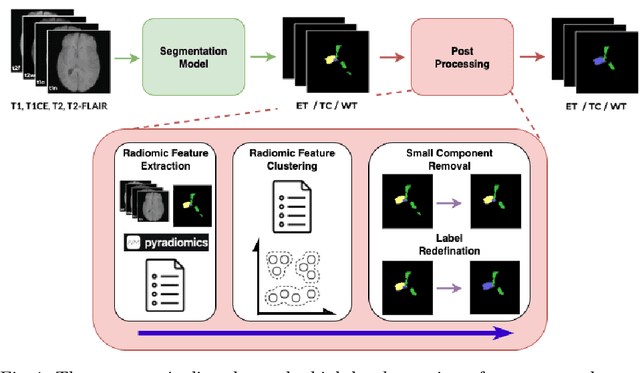
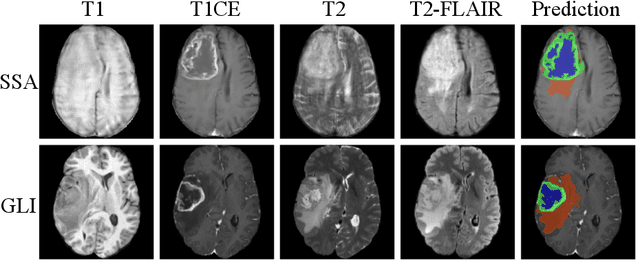

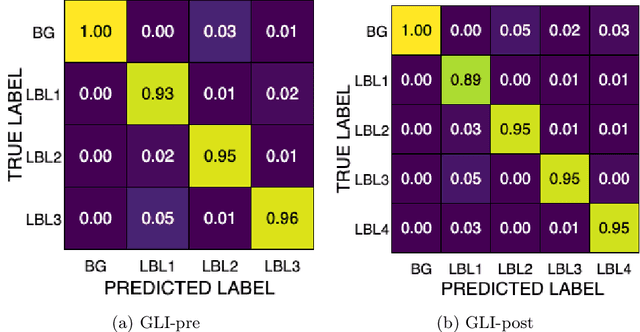
Abstract:Gliomas are the most common malignant brain tumors in adults and are among the most lethal. Despite aggressive treatment, the median survival rate is less than 15 months. Accurate multiparametric MRI (mpMRI) tumor segmentation is critical for surgical planning, radiotherapy, and disease monitoring. While deep learning models have improved the accuracy of automated segmentation, large-scale pre-trained models generalize poorly and often underperform, producing systematic errors such as false positives, label swaps, and slice discontinuities in slices. These limitations are further compounded by unequal access to GPU resources and the growing environmental cost of large-scale model training. In this work, we propose adaptive post-processing techniques to refine the quality of glioma segmentations produced by large-scale pretrained models developed for various types of tumors. We demonstrated the techniques in multiple BraTS 2025 segmentation challenge tasks, with the ranking metric improving by 14.9 % for the sub-Saharan Africa challenge and 0.9% for the adult glioma challenge. This approach promotes a shift in brain tumor segmentation research from increasingly complex model architectures to efficient, clinically aligned post-processing strategies that are precise, computationally fair, and sustainable.
Mechanistic Learning with Guided Diffusion Models to Predict Spatio-Temporal Brain Tumor Growth
Sep 11, 2025Abstract:Predicting the spatio-temporal progression of brain tumors is essential for guiding clinical decisions in neuro-oncology. We propose a hybrid mechanistic learning framework that combines a mathematical tumor growth model with a guided denoising diffusion implicit model (DDIM) to synthesize anatomically feasible future MRIs from preceding scans. The mechanistic model, formulated as a system of ordinary differential equations, captures temporal tumor dynamics including radiotherapy effects and estimates future tumor burden. These estimates condition a gradient-guided DDIM, enabling image synthesis that aligns with both predicted growth and patient anatomy. We train our model on the BraTS adult and pediatric glioma datasets and evaluate on 60 axial slices of in-house longitudinal pediatric diffuse midline glioma (DMG) cases. Our framework generates realistic follow-up scans based on spatial similarity metrics. It also introduces tumor growth probability maps, which capture both clinically relevant extent and directionality of tumor growth as shown by 95th percentile Hausdorff Distance. The method enables biologically informed image generation in data-limited scenarios, offering generative-space-time predictions that account for mechanistic priors.
Analysis of the MICCAI Brain Tumor Segmentation -- Metastases (BraTS-METS) 2025 Lighthouse Challenge: Brain Metastasis Segmentation on Pre- and Post-treatment MRI
Apr 16, 2025Abstract:Despite continuous advancements in cancer treatment, brain metastatic disease remains a significant complication of primary cancer and is associated with an unfavorable prognosis. One approach for improving diagnosis, management, and outcomes is to implement algorithms based on artificial intelligence for the automated segmentation of both pre- and post-treatment MRI brain images. Such algorithms rely on volumetric criteria for lesion identification and treatment response assessment, which are still not available in clinical practice. Therefore, it is critical to establish tools for rapid volumetric segmentations methods that can be translated to clinical practice and that are trained on high quality annotated data. The BraTS-METS 2025 Lighthouse Challenge aims to address this critical need by establishing inter-rater and intra-rater variability in dataset annotation by generating high quality annotated datasets from four individual instances of segmentation by neuroradiologists while being recorded on video (two instances doing "from scratch" and two instances after AI pre-segmentation). This high-quality annotated dataset will be used for testing phase in 2025 Lighthouse challenge and will be publicly released at the completion of the challenge. The 2025 Lighthouse challenge will also release the 2023 and 2024 segmented datasets that were annotated using an established pipeline of pre-segmentation, student annotation, two neuroradiologists checking, and one neuroradiologist finalizing the process. It builds upon its previous edition by including post-treatment cases in the dataset. Using these high-quality annotated datasets, the 2025 Lighthouse challenge plans to test benchmark algorithms for automated segmentation of pre-and post-treatment brain metastases (BM), trained on diverse and multi-institutional datasets of MRI images obtained from patients with brain metastases.
Magnetic Resonance Imaging Feature-Based Subtyping and Model Ensemble for Enhanced Brain Tumor Segmentation
Dec 05, 2024



Abstract:Accurate and automatic segmentation of brain tumors in multi-parametric magnetic resonance imaging (mpMRI) is essential for quantitative measurements, which play an increasingly important role in clinical diagnosis and prognosis. The International Brain Tumor Segmentation (BraTS) Challenge 2024 offers a unique benchmarking opportunity, including various types of brain tumors in both adult and pediatric populations, such as pediatric brain tumors (PED), meningiomas (MEN-RT) and brain metastases (MET), among others. Compared to previous editions, BraTS 2024 has implemented changes to substantially increase clinical relevance, such as refined tumor regions for evaluation. We propose a deep learning-based ensemble approach that integrates state-of-the-art segmentation models. Additionally, we introduce innovative, adaptive pre- and post-processing techniques that employ MRI-based radiomic analyses to differentiate tumor subtypes. Given the heterogeneous nature of the tumors present in the BraTS datasets, this approach enhances the precision and generalizability of segmentation models. On the final testing sets, our method achieved mean lesion-wise Dice similarity coefficients of 0.926, 0.801, and 0.688 for the whole tumor in PED, MEN-RT, and MET, respectively. These results demonstrate the effectiveness of our approach in improving segmentation performance and generalizability for various brain tumor types.
Adult Glioma Segmentation in Sub-Saharan Africa using Transfer Learning on Stratified Finetuning Data
Dec 05, 2024



Abstract:Gliomas, a kind of brain tumor characterized by high mortality, present substantial diagnostic challenges in low- and middle-income countries, particularly in Sub-Saharan Africa. This paper introduces a novel approach to glioma segmentation using transfer learning to address challenges in resource-limited regions with minimal and low-quality MRI data. We leverage pre-trained deep learning models, nnU-Net and MedNeXt, and apply a stratified fine-tuning strategy using the BraTS2023-Adult-Glioma and BraTS-Africa datasets. Our method exploits radiomic analysis to create stratified training folds, model training on a large brain tumor dataset, and transfer learning to the Sub-Saharan context. A weighted model ensembling strategy and adaptive post-processing are employed to enhance segmentation accuracy. The evaluation of our proposed method on unseen validation cases on the BraTS-Africa 2024 task resulted in lesion-wise mean Dice scores of 0.870, 0.865, and 0.926, for enhancing tumor, tumor core, and whole tumor regions and was ranked first for the challenge. Our approach highlights the ability of integrated machine-learning techniques to bridge the gap between the medical imaging capabilities of resource-limited countries and established developed regions. By tailoring our methods to a target population's specific needs and constraints, we aim to enhance diagnostic capabilities in isolated environments. Our findings underscore the importance of approaches like local data integration and stratification refinement to address healthcare disparities, ensure practical applicability, and enhance impact.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge